Why These Python Coders are Joining the napari Community
Python viewer brings together deep learning community and lab scientists
Datasets in science are getting big and unruly. Thankfully, there are people like Carsen Stringer to wrangle them into shape.
A few years ago, she started looking for a way to process terabytes of brain images collected across HHMI’s Janelia Research Campus, where she’s a group leader. Stringer wanted a tool to automatically label different types of neurons. But out-of-the-box products struggled; they had to be retrained for each kind of cell being studied.
So she created Cellpose. Coded in Python, this deep learning-based algorithm segments images into cells using a technique called flow representation.
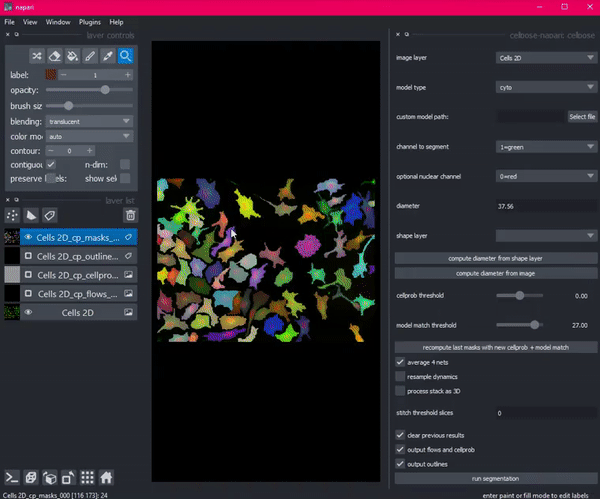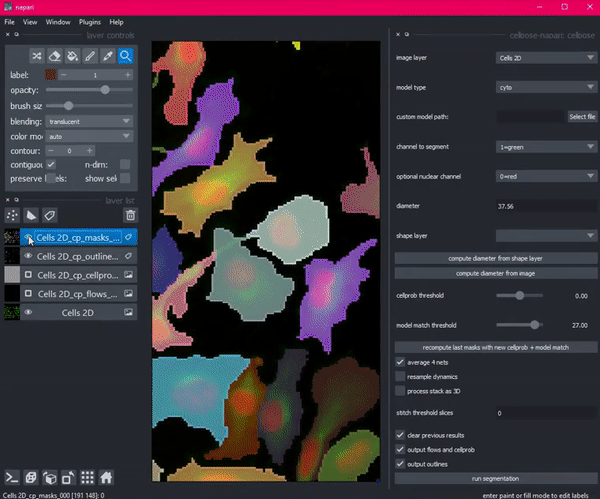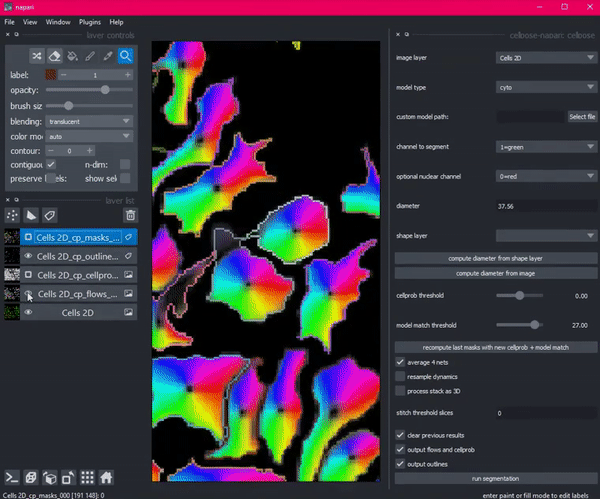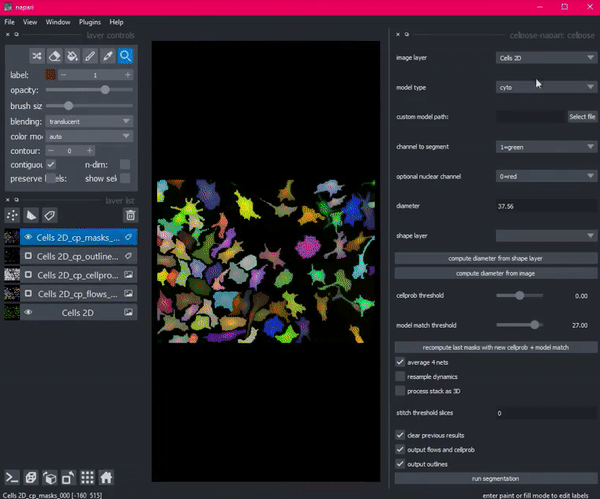
Then Stringer hit a roadblock. She needed a way to see her algorithm in action: a graphical user interface that would allow her research technician to visualize, check and adjust the output of Cellpose. But existing interfaces with this functionality would have required her to port her code from Python to an entirely different language, Java.
“There wasn’t a fully Pythonic GUI that allowed me to label cells and run my code,” said Stringer. “Porting the logic of my code into Java would have been a lot of work.”
More than four thousand miles away in Dresden, Germany, a graduate student named Martin Weigert ran into the same issue while working with data from light-sheet microscopes. He, too, was coding in Python, which has become popular for deep learning, thanks to a wealth of ready-made tools developed in recent years: software libraries like Google’s TensorFlow and Facebook’s PyTorch.
“There is this huge ecosystem of amazing open-source image analysis tools in Python,” said Weigert, now a group leader at the École Polytechnique Fédérale de Lausanne. “But there’s no scientific image viewer to easily take advantage of these tools.”
Stringer and Weigert each ended up building their own interfaces. These homemade platforms worked but required time and effort, and weren’t ideal for sharing their deep-learning methods.
That’s why both researchers have now joined a group of programmers and scientists working on a better solution: napari, a multi-dimensional viewer and image analysis platform coded in Python. Named for an island in the middle of the Pacific Ocean — and supported by an enthusiastic, growing community — napari promises to provide a nexus for Python coders working in science, while making it easier for scientists with limited coding experience to visualize their ever-growing data sets.
“There’s a big need in this space for a Python image viewer,” said Stringer. “Napari is filling that gap.”
A Bridge Between Biologists and Programmers
It’s one thing to build a tool. It’s quite another to convince people to use it and support them when they do.
How many great pieces of code created in a lab have been lost because the student or postdoc behind them moved on to another position? From writing documentation to distributing software to ensuring it runs properly on someone else’s machine, the amount of work required to turn a personal project into something broadly useful can be substantial — and beyond the scope of many researchers’ work.
Scientist Jianxu Chen knows firsthand about this investment in time and resources. He creates Python packages for biologists across the Allen Institute for Cell Science in Seattle, Washington. His team’s Allen Cell and Structure Segmenter breaks down the parts of a cell to visualize them, from the spaghetti-ish mitochondria to the thin fibrous shells of lamin B1 protein encasing a nucleus.

The 29 workflows they have developed are only useful if the workflows are actually easy enough for people to use.
“We are always searching for new ways to make our packages more user-friendly,” said Chen.
Chen experimented with lookup tables and Jupyter notebooks to provide documentation for users. Scientists could load in their data, adapt the workflow to their data and, finally, copy and paste code of the customized workflow into a Python template to process a large batch of images.
Not everyone was familiar with this software, though. Even cutting and pasting code can prove challenging for biologists with little programming experience. So Chen started talking to people about napari.
Chen found its dashboard to be sleek and intuitive. He met with the coders as they integrated on napari’s interface with other scientists to make it more accessible. He was also excited by how it helps users curate results, to create more training data that can further refine deep learning methods.
“Napari creates a bridge between biologists who never look at code and coders who don’t understand biology,” said Chen. “It gives the biologists an intuitive interface to use and the computer scientists a platform that can release their creativity.”
Plugging in with the Alfa Cohort
Adam Tyson agrees. A neuroscientist who studies mouse brains, he has been developing a deep learning napari plugin called cellfinder, which locates individual cells in a whole 3D brain.

Tweaking Cellfinder’s analysis originally required a command-line tool. That proved to be a complicated and iterative process for users. Napari offered easier alternatives, from intuitive buttons and sliders that adjust algorithmic parameters, to its access to backend code. Tyson found its annotations and layers to be useful and its primitives to work nicely for biological data.
Now his team is creating a second plugin for napari that identifies different regions in the brain.
“By bringing this plugin into napari, I hope that people at our institute and across the world working on mouse brains will be able to use it,” said Tyson, a research fellow at UCL’s Sainsbury Wellcome Centre. “My ultimate aim is to find people who want to do similar analyses of large images in other fields, such as developmental biology.”
Chen and Tyson are part of napari’s Alfa Cohort, a group of leading Python image analysis developers that the Chan Zuckerberg Initiative (CZI) Imaging team assembled early this year to work together and brainstorm opportunities for the napari ecosystem while building napari plugins for their tools. The cohort comprised eight project teams in total, and they recently convened virtually to showcase their work. Cellpose and StarDist, which specializes in round objects, were on display, as were Ilastik, created by The European Molecular Biology Laboratory, and YAPiC, by the German Center for Neurodegenerative Diseases, which both outline cells based on pixels. The European Bioinformatics Institute’s SplineIt offers vector representations of object outlines, while nucleAlzer, from the Hungarian Academy of Sciences’ Biological Research Center, focuses on finding nuclei.

The home for all these projects — and future plugins — is the napari hub, a central repository meant to help people discover and distribute bioimage analysis content. Much of these efforts have been made possible by financial and software development support from CZI. CZI also launched a funding opportunity, the napari Plugin Accelerator Grants RFA, to drive development of napari’s growing ecosystem of plugins for image analysis and support the maintenance of plugins.
“Projects like napari are typically hard to get funding for and are squeezed into research grants,” said Uwe Schmidt, an independent researcher who developed StarDist with Weigert. “Having CZI support provides an opportunity to make this large pool of existing scientific software written in Python more easily accessible to people who aren’t comfortable with fiddling with the command line and complicated installation instructions.”
The Open Source Heart of Napari
Getting all of this to work relies on the open-source community coding napari itself, which has its origins in a visit scikit-image developer Juan Nunez-Iglesias paid to San Francisco three years ago. He and Loic Royer, a researcher at the Chan Zuckerberg Biohub, started batting around the idea of a Python viewer. They coded the first version in an afternoon and recruited an undergraduate intern, Kira Evans, to spend a summer expanding it. Then CZI’s Nick Sofroniew joined in, spurring the project onward.
From the beginning, Nunez-Iglesias wanted napari to be n-dimensional: not limited to a few dimensions of space and time, but able to handle an arbitrary number of dimensions.
“In biology you’ll often see a lot of 3D or 3D plus time tools, but you don’t see a lot that are n-dimensional,” said Nunez-Iglesias, a research fellow at the Monash Biomedicine Discovery Institute. “The fact that it is n-dimensional makes it broadly applicable.”
One of his students, Draga Doncila Pop, has already begun to push napari beyond biology. She recently developed a tool to analyze multi-dimensional satellite imagery, which faces many of the same issues as biological imaging.
Supported by an open-source community of coders, napari has continued to grow. Considering napari’s intended use for large datasets, performance and efficiency are key. That’s why Nunez-Iglesias and others are working on making it faster and smoother.
For Talley Lambert of Harvard Medical School, who ranks among those who have contributed significant amounts of code to the project, one of the most exciting parts of the project is the data model just under napari’s hood. It rolls in abstractions for objects like images, points, and surfaces. He hopes this will provide developers a framework to build on each other’s work. A plugin that allows users to find a particular neuron in a brain, for instance, could potentially be followed by another that then outlines the neighboring cells around that neuron.
This community is responsive to feedback, said Lambert, and adds new features as code changes. With the support of CZI staff, napari offers long-term stability — all while keeping the project open source, driven and controlled by its community of python coders.
“Python is known for being a kooky, friendly, open community that welcomes all, and napari follows in this spirit,” said Lambert. “When I see people posting stuff they’re doing with napari on Twitter, applications that surpass what I anticipated it being used for, it gets me super excited about the future of the project.”
